Project Overview
As part of the Probabilistic Graphical Models and Deep Generative Models course by Pierre Latouche and Pierre-Alexandre Mattei, I implemented Mixture Density Networks (MDNs) to model multi-modal regression problems with Antoine Debouchage and Valentin Denée. MDNs allow neural networks to predict full conditional probability distributions rather than single-point estimates, providing richer uncertainty quantification.
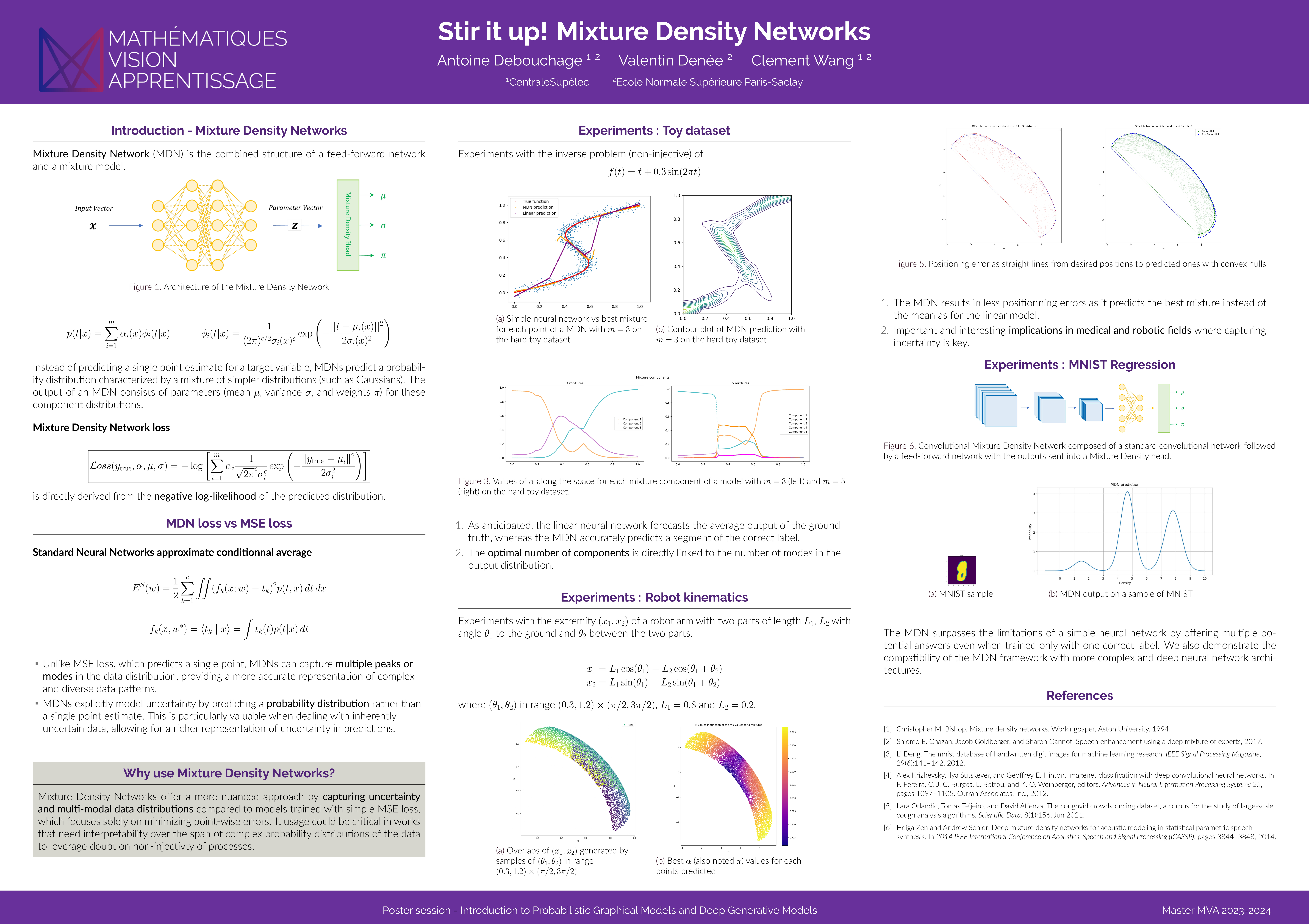
Core concepts
Mixture Density Networks combine neural networks with mixture models. For an input (x), the network predicts the parameters of a mixture of (m) Gaussian components:
\[p(t|x) = \sum_{i=1}^{m} \alpha_i(x) \, \phi_i(t|x)\]- \(\alpha_i(x)\) are the mixing coefficients, interpreted as conditional probabilities that the target \(t\) is generated by the \(i\)-th component.
- \(\phi_i(t\|x)\) are Gaussian kernels with predicted mean \(\mu_i(x)\) and variance \(\sigma_i(x)^2\):
Here, (c) is the dimension of the target vector (t), and we assume the components are independent within each Gaussian.
Unlike standard neural networks that output only a conditional mean (f(x; w)), MDNs provide a full conditional distribution, capturing multiple modes in the data. This is particularly useful in regression tasks where the target is inherently multi-modal.
Key implementation details
- The neural network outputs three sets of parameters per component: mixing coefficients, means, and variances.
- Training is done using maximum likelihood estimation, minimizing the negative log-likelihood of the observed targets under the predicted mixture distribution.
- The weights are updated with Adam optimizer.
- We tested MDNs on several synthetic and real datasets to evaluate their ability to capture multi-modal patterns and uncertainty.
Documentation
For detailed explanations, figures, and results, please consult the following resources:
-
Previous
Satellite Images Competition and Publication at BiDS 2023 -
Next
ABBA Symbolic Representation of Time Series