Project Overview
This project, part of the Advanced Learning for Text and Graph Data course, tackled a challenging task: retrieve the correct molecular structure from a text description.
We combined textual descriptions and molecular graphs using a contrastive learning framework, aligning representations of molecules and their textual descriptions in a shared embedding space.
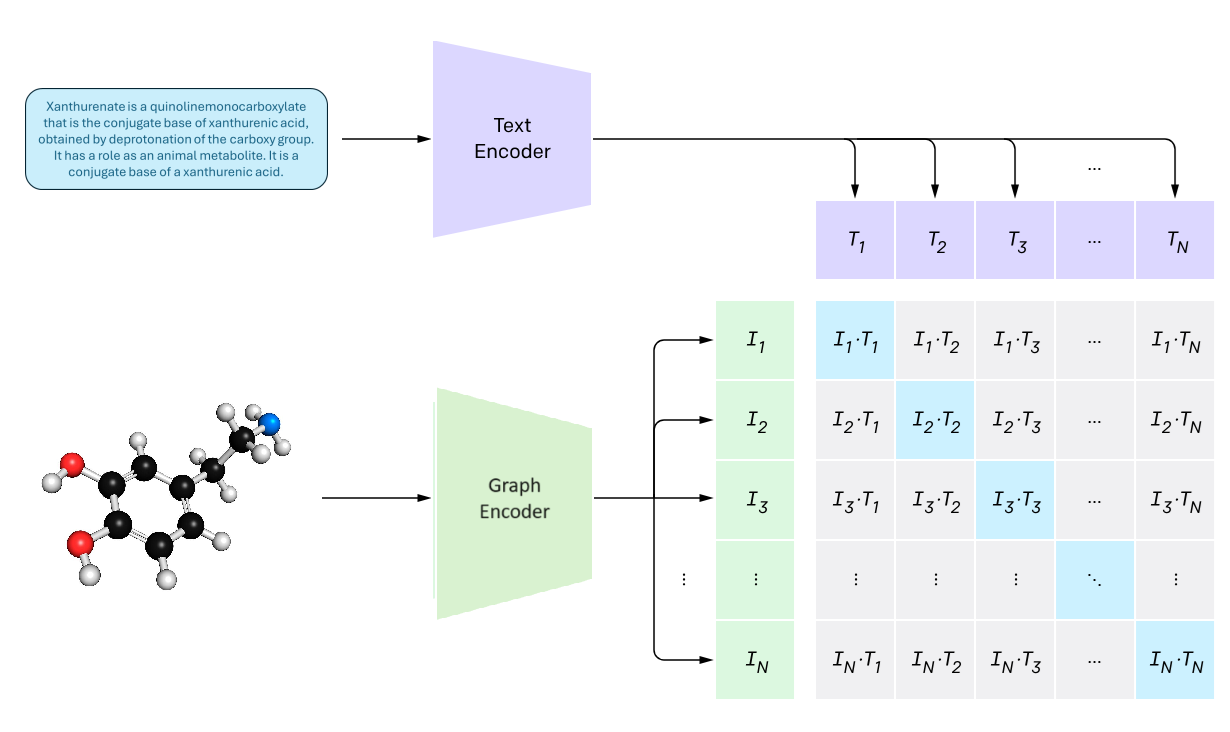
Methodology
Text Encoder
Molecule names and descriptions often contain specialized vocabulary. Standard tokenizers split them poorly, so we trained a custom tokenizer to preserve meaningful units. On top of this, we trained a DistilBERT model from scratch using masked language modeling on the text data from the dataset. This allowed the text encoder to generate embeddings that capture semantic nuances of chemical descriptions.
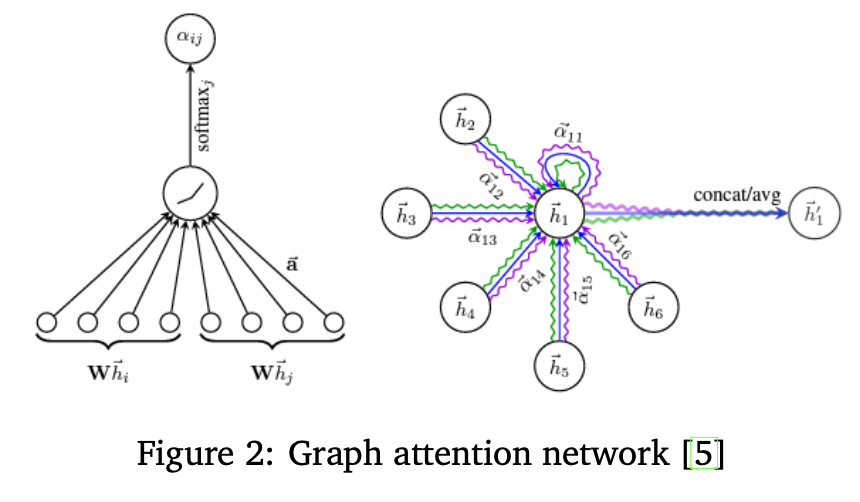
Graph Encoder
Molecules are naturally represented as graphs. We used Graph Attention Networks (GATs) to encode molecule graphs. We chose GAT because of its ability to attribute dynamic importance to neighboring atoms and bonds. The graph encoder outputs embeddings that summarize the molecular graph in a way that can be directly compared to text embeddings.
Training Tricks
Training stability and convergence were critical. We used cosine learning rate scheduling with warmup, large batch sizes, and online hard sample mining. Hard sample mining prioritized the most challenging molecules within a batch, improving contrastive learning effectiveness. Dropout and layer normalization prevented overfitting. We also used mixed-precision training to enable scaling to larger batch sizes. In contrastive learning, having larger batch sizes is crucial to obtain harder negative samples.
Graph Augmentations
To improve generalization, we augmented graphs during training:
- Edge perturbation: random removal or addition of edges.
- Graph sampling and k-hop subgraphs: train on partial views of the molecule.
- Feature masking and noise: hide or perturb node features to encourage robustness.
Edge perturbation had the largest positive impact, while more aggressive augmentations sometimes degraded performance.
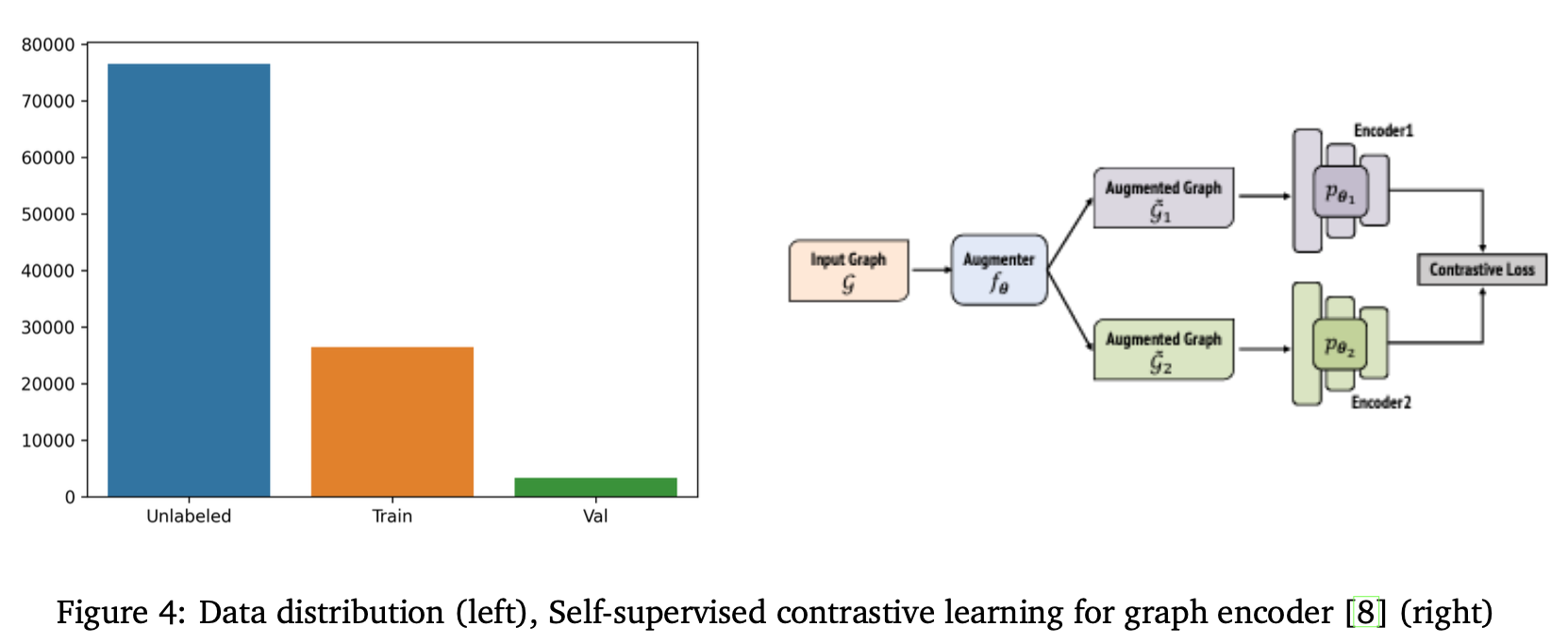
Self-Supervised Training
Labeled data was limited, so we leveraged self-supervised contrastive learning. Inspired by the iBOT framework, we trained a student and teacher network using multiple augmented views of each molecule. The student was optimized to match the teacher’s embeddings for corresponding views, promoting invariant representations. Centering and temperature scaling stabilized training. This pretraining improved convergence and helped the model generalize better on unseen molecules.
Implementation details
We evaluated our approach using a combination of labeled and unlabeled data. The final model was an ensemble of two GAT-based models:
- DistilBERT text encoder fine-tuned on molecule descriptions.
- GAT graph encoder with 6 layers, hidden dimension 512, and 0.2 dropout.
- Cosine learning rate scheduling with warmup, AdamW optimizer, large batch sizes.
- Online hard sample mining and edge perturbation augmentations.
- One model used self-supervised pretraining on unlabeled molecules.
This setup achieved the best Label Ranking Average Precision (LRAP) on the validation set and generalized well to the Kaggle leaderboard.
Documentation
References
- Devlin, J., Chang, M.-W., Lee, K., Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. 2018.
- Sanh, V., Debut, L., Chaumond, J., Wolf, T. DistilBERT: Smaller, Faster, Cheaper and Lighter. 2019.
- Frey, N., Soklaski, R., Axelrod, S., et al. Neural Scaling of Deep Chemical Models. ChemRxiv, 2022.
- Lee, J., Yoon, W., Kim, S., et al. BioBERT: Pre-trained Biomedical Language Representation Model. 2020.
- Veličković, P., Cucurull, G., Casanova, A., et al. Graph Attention Networks. 2017.
- Gao, L., Zhang, Y. Scaling Deep Contrastive Learning Batch Size. 2021.
- Shrivastava, A., Gupta, A., Girshick, R. Online Hard Example Mining. 2016.
- Ding, K., Xu, Z., Tong, H., Liu, H. Data Augmentation for Deep Graph Learning: A Survey. 2022.
- Zhou, J., Wei, C., Wang, H., et al. iBOT: Image BERT Pre-training with Online Tokenizer. 2021.
- Rendle, S., Freudenthaler, C., Gantner, Z., Schmidt-Thieme, L. BPR: Bayesian Personalized Ranking from Implicit Feedback. 2012.
-
Previous
ABBA Symbolic Representation of Time Series -
Next
Implicit Acceleration by Overparameterization